
Mucosal
Crosstalk
laboratory
Laboratory for Mucosal Crosstalk
We are the laboratory for Mucosal Crosstalk. Our research is dedicated to understanding how the interactions between our microbiota, the intestinal epithelium, and immune cells in the intestinal mucosa maintain intestinal homeostasis, and how these interactions derail in disease. To achieve this, we employ interdisciplinary approaches at the interface of epithelial cell biology, microbiology and mucosal immunology in combination with versatile in vivo and in vitro models.
The intestinal mucosa forms an integral barrier between our bodies and the outside world. Being exposed to a huge variety of metabolites and microbes taken up with our food or stably colonizing the intestine, it is permeable for essential nutrients while serving as a protective barrier to prevent microbial translocation. A tight balance between the intestinal epithelium, immune cells in the underlying tissue and the intestinal microbiota present in the lumen maintains intestinal homeostasis. The impairment of this balance can trigger disease, such as infection, metabolic disorders, and chronic inflammation (e.g. inflammatory bowel disease). Our work focuses on understanding how these key players interact to maintain intestinal homeostasis, with a special interest in the intestinal epithelium. The intestinal epithelium is in direct contact with the microbiota colonizing the intestinal lumen (e.g. sensing via pattern recognition receptors, uptake of metabolites), yet we know little about the molecular basis of how the epithelium integrates luminal and mucosal stimuli. We make use of state-of-the-art in vivo and advanced organoid-based in vitro models, (single cell) omics techniques and bioengineering approaches, to map mucosal cellular crosstalk and dissect the underlying molecular mechanisms in mouse and human.
Our Research
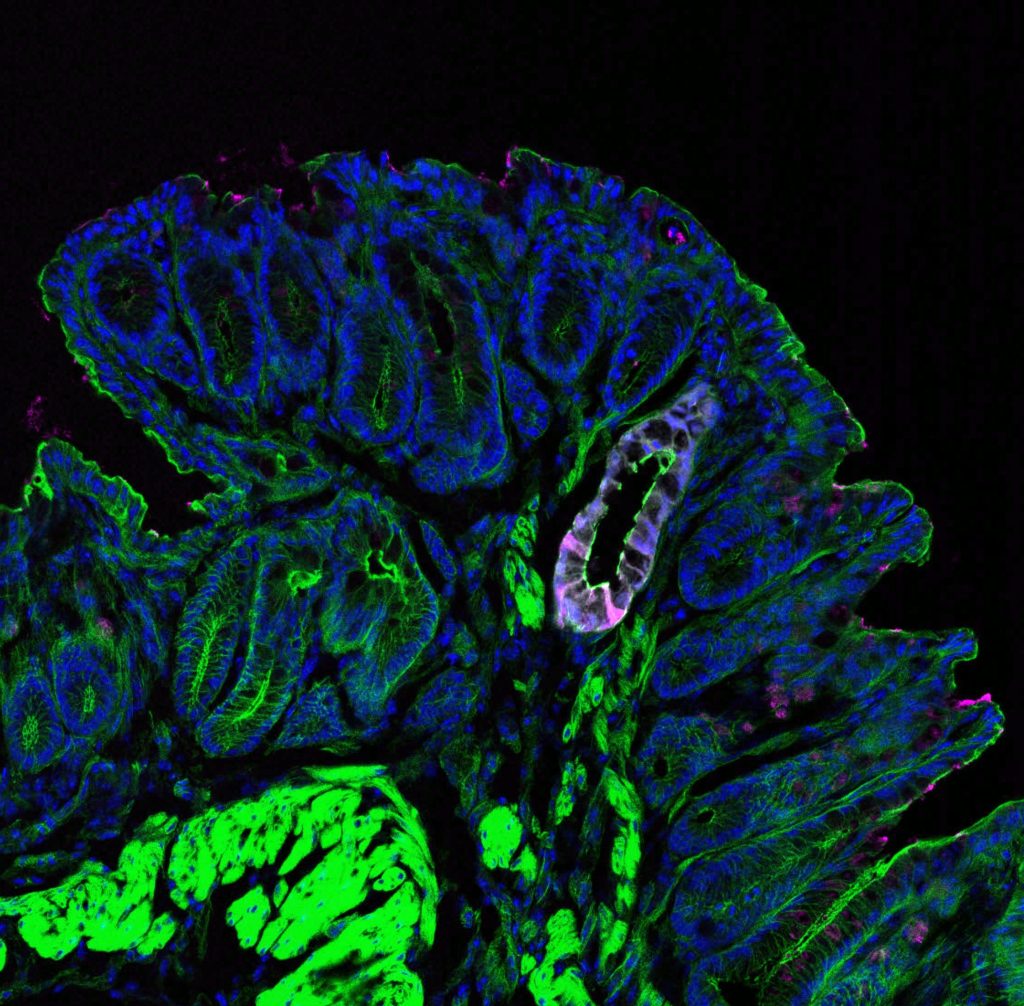
Characterization of microbiota impact on epithelial phenotypes
Using state-of-the-art in vivo and in vitro models, we characterize the impact of commensal bacteria on epithelial phenotypes using (sc) omics, microscopy and flow cytometry.
In vivo models
We employ gnotobiotic mice associated with microbial consortia of varying complexity to mechanistically dissect the impact of commensal bacteria on the intestinal epithelium. Using orthotopic transplantation, we can transplant genetically modified organoids into the murine colon.
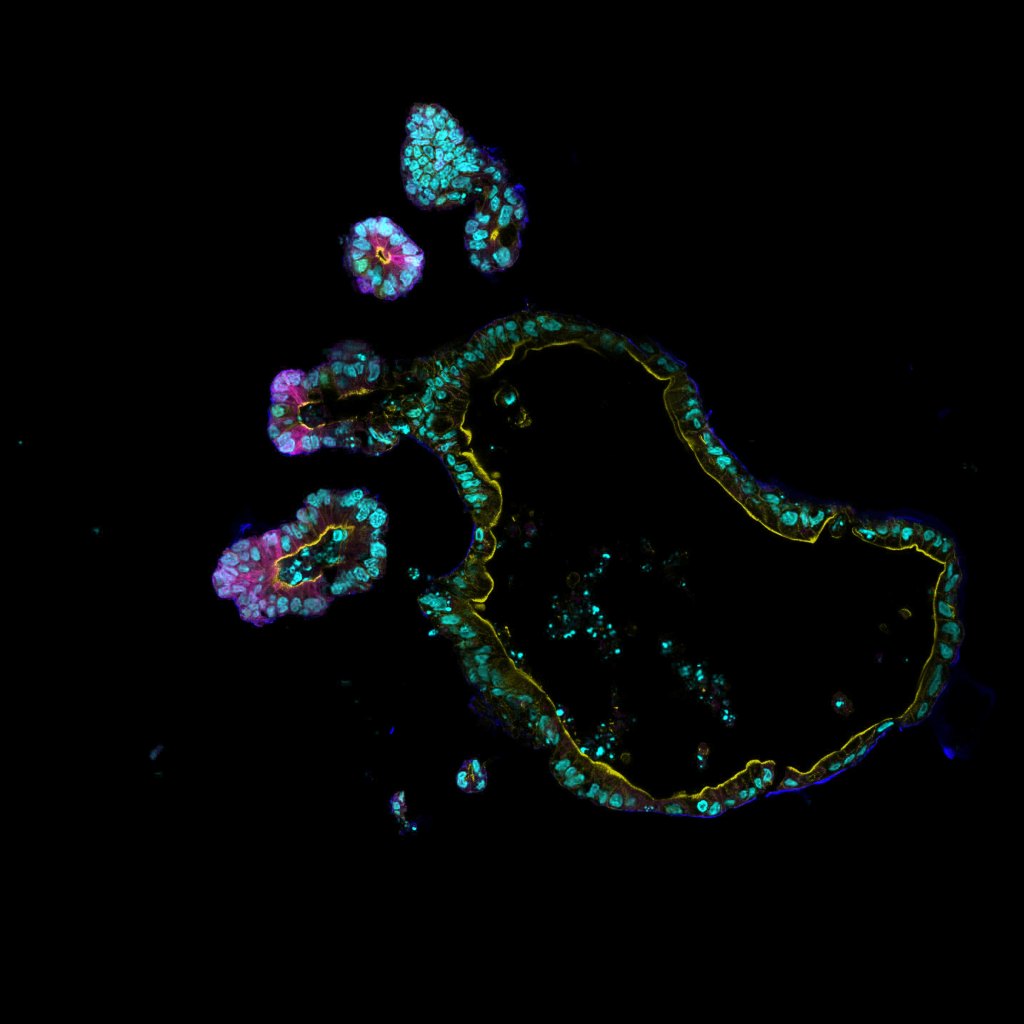
In vitro models
We use human and murine organoids in combination with cultivation of bacterial consortia and bioengineering approaches to develop novel in vitro model systems for the molecular dissection of epithelium-microbiota interactions.










